1) Next Generation Sequencing (NGS) - An Introduction
Summary
TLDRThis video provides an in-depth overview of Next Generation Sequencing (NGS) technologies, exploring their significance in fields like personalized medicine, genetics, and diagnostics. It contrasts NGS with Sanger Sequencing, highlighting differences in methodology and applications. The video covers four primary NGS techniques—pyrosequencing, sequencing by synthesis, sequencing by ligation, and ion semiconductor sequencing—detailing their unique processes, advantages, and limitations. Additionally, it discusses how these technologies impact genome sequencing coverage, with a focus on the efficiency of modern platforms. Viewers are encouraged to explore various NGS services offered by abm for advanced sequencing solutions.
Takeaways
- 😀 Next Generation Sequencing (NGS) enables simultaneous sequencing of thousands to millions of DNA molecules, revolutionizing personalized medicine, genetic diseases, and clinical diagnostics.
- 😀 Sanger Sequencing, the gold standard developed in the 1900s, is still used extensively for routine DNA sequencing and validating NGS data.
- 😀 The Sanger Sequencing method involves using di-deoxynucleotides that terminate DNA chains at specific nucleotide positions and allow for automatic sequencing through fluorescent detection.
- 😀 NGS platforms differ from Sanger Sequencing by enabling massively parallel sequencing, capable of analyzing millions or even billions of sequences simultaneously.
- 😀 NGS platforms use four main technologies: pyrosequencing, sequencing by synthesis, sequencing by ligation, and ion semiconductor sequencing.
- 😀 Pyrosequencing detects light emission from pyrophosphate release during nucleotide incorporation and is known for long read lengths but suffers from high error rates in homopolymers.
- 😀 Sequencing by synthesis, employed by Illumina, uses reversible fluorescent terminator nucleotides for single-base incorporation and generates high-quality data, though it faces increasing error rates as the sequencing progresses.
- 😀 Sequencing by ligation uses short 8-mer probes to hybridize and ligate to DNA, generating shorter reads with fewer errors but limited by the short length of sequences.
- 😀 Ion semiconductor sequencing detects pH changes due to hydrogen ion release during nucleotide incorporation, offering a cost-effective alternative but with a lower read quality compared to other methods.
- 😀 For reliable whole genome sequencing, a minimum of 30x coverage is required, with NGS systems capable of sequencing multiple genomes with high coverage in a single run.
- 😀 The sequencing by synthesis method (e.g., Illumina HiSeq) provides high throughput, capable of sequencing up to 15 individuals in a few days, while Ion Proton machines are often used in clinical settings for rapid results.
Q & A
What is Next Generation Sequencing (NGS), and how is it used in research?
-Next Generation Sequencing (NGS) is a powerful platform that enables the sequencing of thousands to millions of DNA molecules simultaneously. It is used in research to study genetic variations, identify mutations, and analyze genomes in various fields like personalized medicine, genetic disease research, and clinical diagnostics.
How does Sanger Sequencing differ from NGS?
-Sanger Sequencing, developed in the 1900s, is the gold standard for DNA sequencing and is still used for routine applications and validating NGS data. It involves the incorporation of dideoxynucleotides to terminate DNA strand elongation, while NGS is a massively parallel technology capable of sequencing millions of DNA fragments at once.
What role do dideoxynucleotides play in Sanger Sequencing?
-Dideoxynucleotides are modified nucleotides used in Sanger Sequencing that lack a 3' hydroxyl group, preventing further DNA strand elongation and resulting in chain termination. Each dideoxynucleotide is also tagged with a unique fluorescent dye, which enables the detection of DNA sequence during the process.
What are the common features shared by all NGS platforms?
-All NGS platforms have three common features: sample preparation, where DNA libraries are created via amplification or ligation with custom adapters; sequencing machines, which amplify DNA fragments on a solid surface; and data output, where raw data in the form of DNA sequences is generated after sequencing reactions.
What are the key differences between pyrosequencing and sequencing by synthesis?
-Pyrosequencing monitors nucleotide incorporation through light emission triggered by the release of pyrophosphate, while sequencing by synthesis (used by Illumina platforms) relies on the incorporation of reversibly fluorescent and terminated nucleotides. Pyrosequencing offers longer read lengths but has higher costs and error rates with homopolymers, whereas sequencing by synthesis has more accurate reads but faces increasing error rates as sequencing progresses.
What are the advantages and disadvantages of sequencing by ligation?
-Sequencing by ligation uses oligonucleotide probes instead of DNA polymerase, allowing for short reads. It is precise but produces shorter read lengths compared to other NGS methods. The major disadvantage is the limited output, and it requires several cycles to complete sequencing, which can reduce efficiency.
How does ion semiconductor sequencing detect nucleotide incorporation?
-Ion semiconductor sequencing detects nucleotide incorporation by measuring the release of hydrogen ions during DNA synthesis. Each cluster of DNA is positioned above a semiconductor transistor, which detects pH changes caused by the release of hydrogen ions when a nucleotide is incorporated.
What is the minimum coverage required for whole genome sequencing to be valid?
-For whole genome sequencing to be useful, a minimum coverage of 30x is required. This ensures that each part of the genome is sequenced enough times to provide reliable and accurate data.
Which NGS method is most commonly used for large-scale sequencing projects, and why?
-Sequencing by synthesis, particularly using Illumina platforms, is the most commonly used method for large-scale sequencing projects. It can generate hundreds of coverage per run and sequence multiple individuals simultaneously, making it efficient for large-scale studies.
What are some of the NGS services offered by abm, and how can they be accessed?
-abm offers a wide range of NGS services, including whole genome sequencing, exome sequencing, RNA sequencing, and disease panels. These services can be accessed by visiting the abm website, where detailed information about each service, including technical details, pricing, and bioinformatics solutions, is available.
Outlines

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowMindmap

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowKeywords

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowHighlights

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowTranscripts

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowBrowse More Related Video

DNA Sequencing Techniques | An Overview

Genética e Biologia Molecular – Aula 01 – O DNA: uma sinopse histórica

Introduction to Next Generation Sequencing
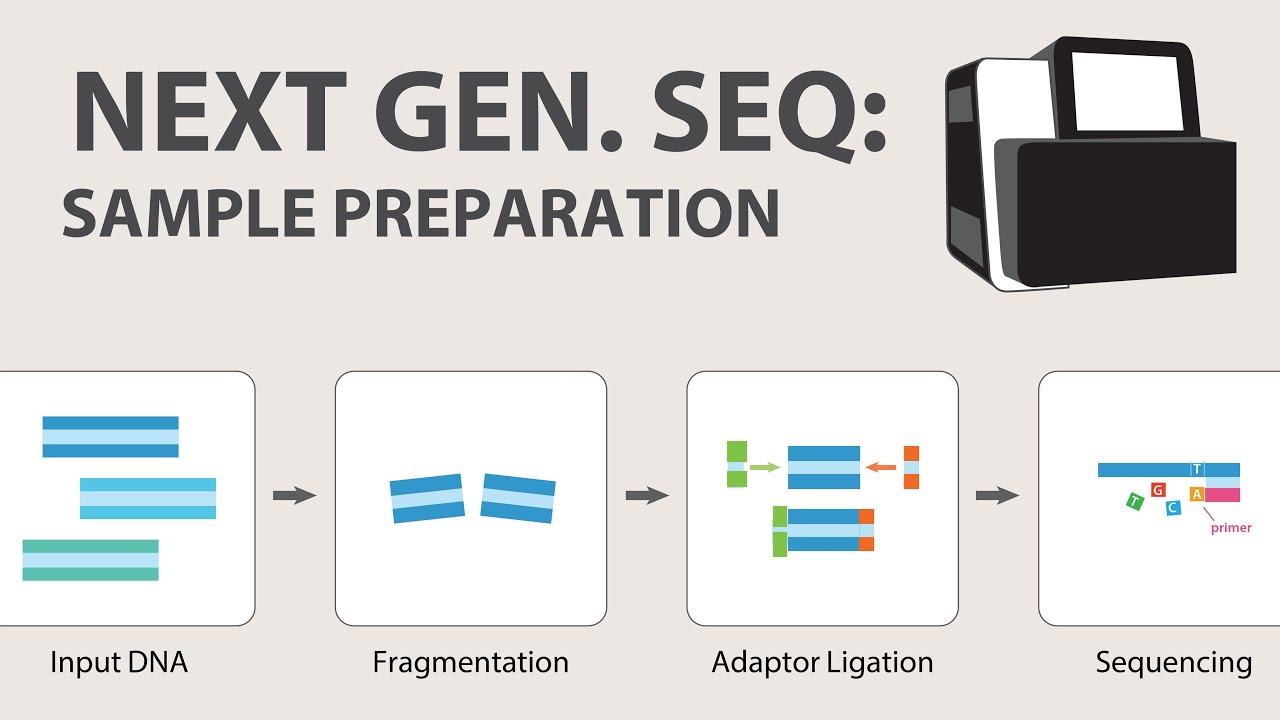
2) Next Generation Sequencing (NGS) - Sample Preparation
Next Generation Sequencing - A Step-By-Step Guide to DNA Sequencing.
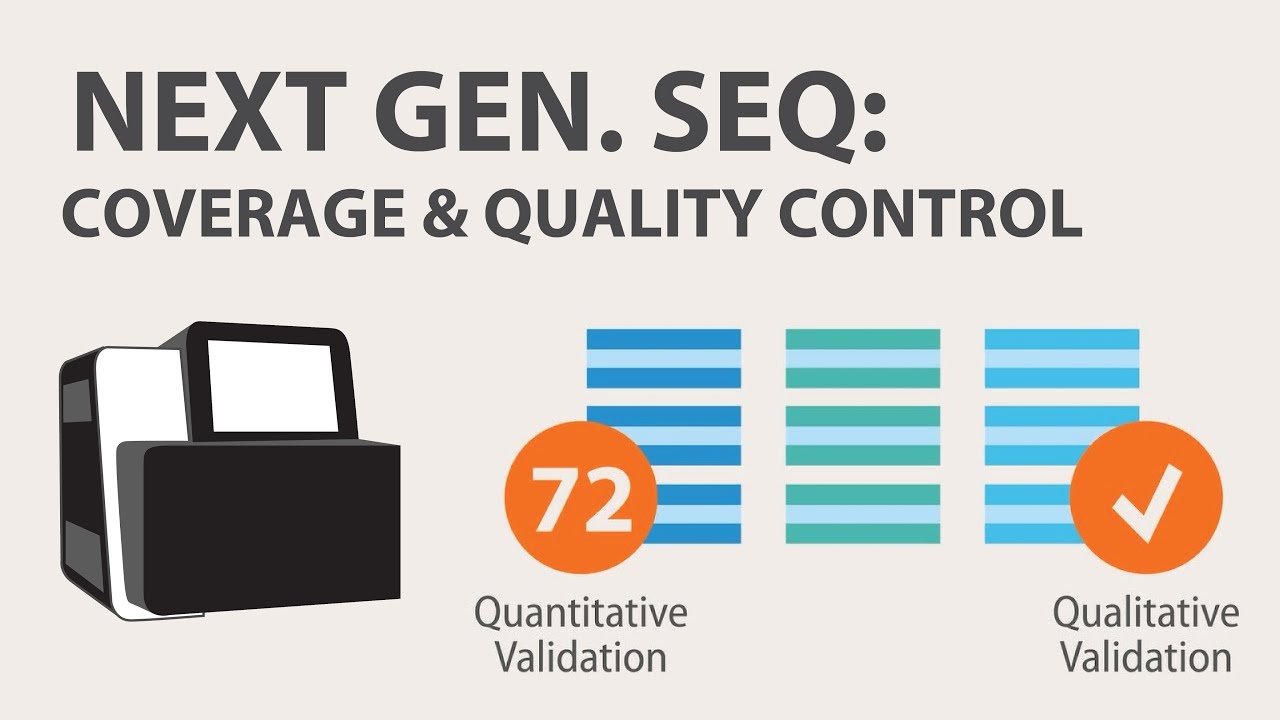
3) Next Generation Sequencing (NGS) - Coverage & Sample Quality Control
5.0 / 5 (0 votes)