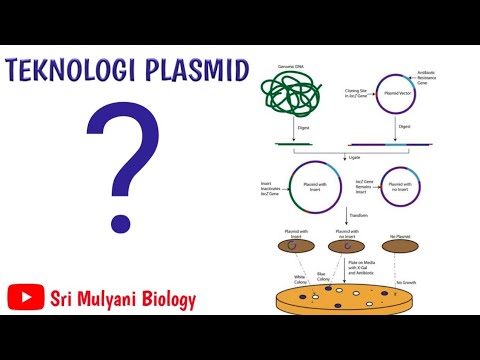
How do Antibiotic Resistance Genes function as selectable marker or helps in transformant selection?
Summary
TLDRThis video explains how antibiotic resistance genes serve as selectable markers in the selection of recombinant colonies. It uses the PPR 322 vector as an example, demonstrating how the ampicillin and tetracycline resistance genes enable the growth of recombinant bacteria. By inserting a gene of interest into the tetracycline resistance region, the gene becomes non-functional through insertional inactivation, allowing for easy selection. The video also covers the use of replica plating to identify colonies with the gene of interest, offering a detailed yet clear explanation of how antibiotic resistance helps in the selection process.
Takeaways
- 😀 A selectable marker is a gene sequence, such as an antibiotic resistance gene, that helps in selecting recombinant colonies.
- 😀 The PPR 322 vector contains two selectable markers: ampicillin resistance and tetracycline resistance genes.
- 😀 Antibiotic resistance genes make bacteria resistant to specific antibiotics, allowing them to grow in media containing those antibiotics.
- 😀 Insertional inactivation occurs when the gene of interest is inserted into a selectable marker gene, making it nonfunctional.
- 😀 Using restriction enzymes, like BamHI, allows insertion of the gene of interest into a selectable marker region.
- 😀 The recombinant vector with the inserted gene of interest will have an inactivated tetracycline resistance gene.
- 😀 After transformation, there are three types of colonies: non-transformed, transformed with non-recombinant vector, and transformed with recombinant vector.
- 😀 Non-transformed colonies cannot grow on antibiotic-containing media because they lack resistance genes.
- 😀 Colonies transformed with a non-recombinant vector can grow on both tetracycline and ampicillin media but don’t contain the gene of interest.
- 😀 Recombinant colonies, with the gene of interest inserted, can grow on ampicillin but not tetracycline media due to tetracycline gene inactivation.
- 😀 Replica plating is used to transfer colonies from a master plate to selective media, helping identify colonies with the recombinant vector.
Q & A
What is a selectable marker in genetic engineering?
-A selectable marker is a gene sequence, like an antibiotic resistance gene, within a vector that helps in selecting recombinant colonies. It ensures that only cells with the recombinant DNA are able to grow in the presence of specific antibiotics.
How does the antibiotic resistance gene help in the selection of recombinant colonies?
-The antibiotic resistance gene allows transformed bacteria to survive in the presence of the corresponding antibiotic. This makes it easier to select colonies that contain the recombinant DNA, as only those with the resistance gene will grow on selective media.
What is the purpose of the PPR 322 vector in the transcript?
-The PPR 322 vector serves as an example of a vector that contains two selectable markers: ampicillin resistance and tetracycline resistance. These markers help in the selection process for recombinant colonies in genetic engineering experiments.
What are the two selectable markers mentioned in the PPR 322 vector?
-The two selectable markers in the PPR 322 vector are the ampicillin resistance gene and the tetracycline resistance gene.
What happens when the gene of interest is inserted into the tetracycline resistance gene?
-Inserting the gene of interest into the tetracycline resistance gene causes 'insertional inactivation,' meaning the tetracycline resistance gene becomes nonfunctional, making it easier to distinguish recombinant colonies.
What are the three types of colonies observed after a transformation experiment?
-The three types of colonies are: 1) Non-transformed colonies (without the vector), 2) Transformed colonies with a non-recombinant vector (no gene of interest), and 3) Transformed colonies with a recombinant vector (with the gene of interest).
What is replica plating, and how is it used in colony selection?
-Replica plating is a technique where an imprint of bacterial colonies is transferred to a new plate to maintain their spatial organization. It is used to transfer colonies to selective media, helping identify those that contain recombinant DNA.
How does replica plating help identify transformed colonies with the gene of interest?
-By using replica plating, colonies that grow on ampicillin-containing medium but not on tetracycline-containing medium are identified as containing the recombinant vector with the gene of interest, as the insertion of the gene disables the tetracycline resistance.
Why can't non-transformed colonies grow on antibiotic-containing media?
-Non-transformed colonies lack the antibiotic resistance genes, so they cannot survive in the presence of antibiotics like tetracycline or ampicillin, which are used to select recombinant colonies.
What is the significance of the non-recombinant vector in the selection process?
-Colonies transformed with a non-recombinant vector can grow on both tetracycline and ampicillin media because both resistance genes are intact, but they do not contain the gene of interest. These colonies are discarded during selection.
Outlines

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowMindmap

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowKeywords

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowHighlights

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowTranscripts

This section is available to paid users only. Please upgrade to access this part.
Upgrade Now5.0 / 5 (0 votes)