Predicting protein function with ProtNLM
Summary
TLDRGoogle researchers collaborated with the European Bioinformatics Institute to develop 'Protein LM,' a model predicting protein functions from amino acid sequences. This breakthrough aids in identifying disease-related proteins and drug targets, potentially revolutionizing drug discovery. With over 150 million proteins in its training data, 'Protein LM' accurately annotates sequences, even those previously uncharacterized. The model not only generates protein names but also retrieves relevant proteins from databases, showcasing the significant impact of machine learning on scientific advancements.
Takeaways
- 🔬 Google Research collaborated with the European Bioinformatics Institute to predict protein functions using a model called Protein LM.
- 🧬 Protein LM predicts short functional descriptions, known as protein names, from amino acid sequences.
- 🌐 The model's predictions are used to label millions of previously unannotated protein sequences in databases.
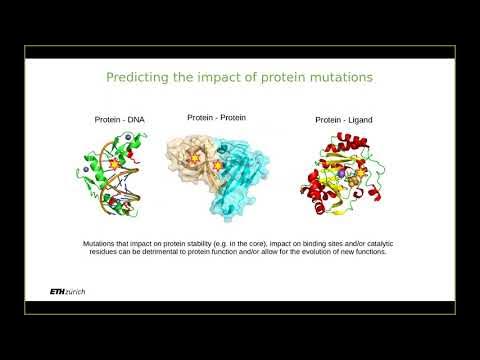
- 💊 Understanding protein roles is crucial for identifying disease connections and potential drug targets.
- 🔎 Developing new proteins for drug design is a complex and expensive process that involves significant trial and error.
- 🧬 Each protein is composed of a sequence of amino acids, which determine its function, but decoding this sequence is challenging.
- 🔍 The Protein LM model uses natural language processing to translate amino acid sequences into functional descriptions.
- 📈 The model faced challenges in training and evaluation due to the vast amount of data and the complexity of protein sequences.
- 📊 The model's accuracy was validated through automated evaluation, showing high percentages of correct predictions.
- 🔧 The model can be used to generate descriptions for simple proteins or to discover relevant proteins in databases based on descriptions.
- 🌟 Google Research released annotations for millions of previously unnamed full-length protein sequences and provided tools for users to experiment with the models.
Q & A
What is the primary goal of the collaboration between Google Research and the European Bioinformatics Institute?
-The primary goal is to predict the function of proteins by developing a model called Protein LM, which can predict a short functional description, or protein name, from the protein's amino acid sequence.
What is Protein LM and how does it work?
-Protein LM is a model developed to predict the function of proteins. It works by taking an amino acid sequence as input and generating a short natural language description of the protein's function, similar to generating a protein name.
How does understanding protein function contribute to scientific discoveries?
-Understanding protein function is critical for identifying proteins connected to diseases, which can be used as drug targets. It also enables the design of new proteins, leading to advances in drug discovery and potentially curing diseases like Ebola.
What challenges does the current process of protein annotation face?
-The current process faces challenges such as the slow and expensive nature of experimental annotation, and the fact that less than one percent of naturally occurring protein sequences have been experimentally characterized.
How does the Protein LM model address the challenge of protein annotation?
-Protein LM addresses this challenge by leveraging advancements in language models and available protein sequences to predict natural language descriptions of amino acid sequences, thus annotating proteins that previously had no description.
What is the significance of the model's ability to generate new names for proteins?
-The model's ability to generate new names for proteins is significant because it allows for the annotation of proteins that have no existing names or descriptions, expanding the database of known protein functions.
How does the model handle the complexity of full protein sequences?
-The model handles the complexity by taking the amino acid sequence input in the form of a sequence of characters, one amino acid at a time, and outputs the name or description one token at a time.
What are the challenges in evaluating the accuracy of protein function predictions?
-Evaluating the accuracy of protein function predictions is challenging because natural language descriptions are hard to evaluate, and there is no visual way to detect correctness. Additionally, existing information and bioinformatics tools may not always prove or disprove the correctness of a prediction.
How does the team plan to improve the Protein LM model in the future?
-The team plans to improve the model by incorporating feedback from manual and automatic evaluations, user feedback, and adding new sources of information as input when available.
What are some future directions the research team is looking forward to exploring?
-The research team is looking forward to exploring tasks like predicting longer functional descriptions, and the inverse problem of generating a new protein sequence from a user-defined description.
How does the collaboration with the European Bioinformatics Institute benefit the project?
-The collaboration allows for the opportunity to improve the models with each UniProt release, ensuring that the annotations remain up-to-date and accurate.
Outlines

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowMindmap

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowKeywords

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowHighlights

This section is available to paid users only. Please upgrade to access this part.
Upgrade NowTranscripts

This section is available to paid users only. Please upgrade to access this part.
Upgrade Now5.0 / 5 (0 votes)