1) PCR (Polymerase Chain Reaction) Tutorial - An Introduction
Summary
TLDRThis video from Applied Biological Materials introduces PCR, a vital molecular biology technique used for DNA amplification in various applications like sequencing and diagnostics. It explains the PCR process, including denaturation, annealing, and elongation, and highlights the importance of factors like primer design and DNA polymerase choice. The video also discusses challenges and solutions, such as GC-rich templates and PCR inhibitors, and promotes abm's range of PCR products.
Takeaways
- 🔬 PCR, or Polymerase Chain Reaction, is a fundamental technique in genetic and molecular biology with applications in DNA sequencing, forensics, and disease diagnosis.
- 📈 PCR enables the rapid and cost-effective amplification of DNA fragments from a small quantity of nucleic acid, often referred to as 'molecular photocopying'.
- 🔄 The PCR process involves repeated cycles of DNA replication, doubling the number of DNA strands with each cycle, resulting in over a trillion copies from a single DNA molecule.
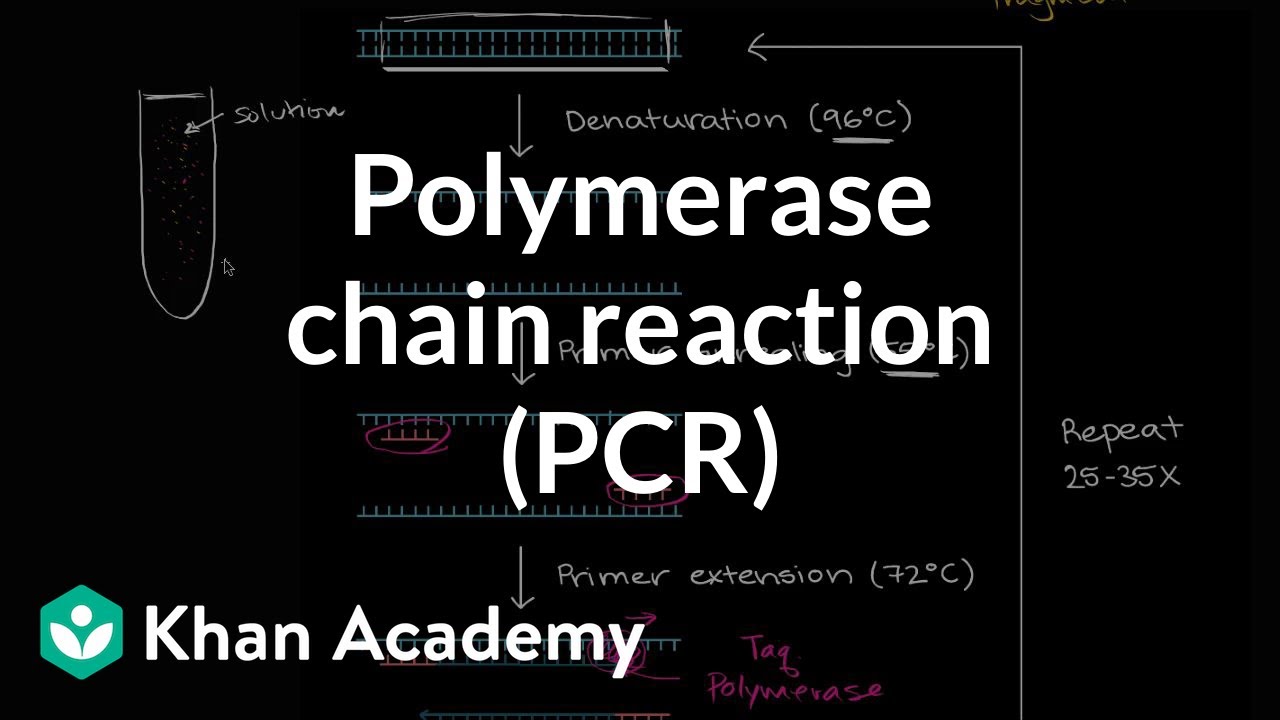
- 🔥 The PCR procedure consists of four main steps: initialization, denaturation, annealing, and elongation, each with specific temperature requirements.
- 🔬 Initialization involves heating to activate DNA polymerase and denature contaminants, facilitating cell lysis in the case of colony PCR screening.
- 🧬 Denaturation breaks the hydrogen bonds of double-stranded DNA by heating, separating the strands for primer binding and replication.
- 🔍 Annealing lowers the temperature to allow primers to bind to the DNA template, guiding the DNA polymerase for replication.
- 📝 Elongation involves the synthesis of a new DNA strand by DNA polymerase incorporating dNTPs, with conditions depending on the enzyme and target sequence.
- 🔥 Taq Polymerase is the most commonly used enzyme in PCR, being thermostable and allowing for multiple amplification cycles without the need for enzyme replenishment.
- 🛠️ Variations of Taq enzyme have been engineered for specific PCR applications, such as those with decreased error rates or the ability to amplify DNA directly from blood samples.
- 🧪 Gel electrophoresis is used to visualize and estimate the size of amplified DNA fragments, ensuring successful gene amplification.
- 🚫 Many factors can interfere with PCR, including DNA template composition, enzyme choice, buffer components, primer design, and the presence of inhibitors.
Q & A
What is Polymerase Chain Reaction (PCR)?
-PCR is a molecular biology technique that allows for the fast and inexpensive amplification of DNA fragments, generating large quantities of DNA from a small amount of nucleic acid, and is often referred to as molecular photocopying.
What are some common applications of PCR?
-PCR is widely used in DNA sequencing, DNA fingerprinting, forensics, detection of microorganisms, and diagnosis of hereditary diseases.
How does PCR work in terms of DNA replication cycles?
-PCR depends on 20 to 40 repeated cycles of DNA replication by a DNA polymerase enzyme, doubling the number of DNA strands after each cycle, resulting in over a trillion copies from a single DNA molecule.
What are the four main steps of the PCR process?
-The PCR process consists of initialization, denaturation, annealing, and elongation.
What happens during the initialization step of PCR?
-In the initialization step, the reaction is heated to activate the DNA polymerase and to denature other contaminants, facilitating cell lysis to release DNA and denature cellular proteins in the case of colony PCR screening.
What is the purpose of the denaturation step in PCR?
-The denaturation step involves breaking hydrogen bonds between the double-stranded DNA by heating, which separates the DNA strands to prepare for primer binding.
What role do primers play in the annealing step of PCR?
-Primers bind to a complementary sequence in the DNA template during the annealing step, guiding the DNA polymerase enzyme for replication.
What is the function of DNA polymerase during the elongation step?
-DNA polymerase incorporates dNTPs in a 5' to 3' direction to synthesize a new DNA strand during the elongation step.
Why is Taq polymerase commonly used in PCR?
-Taq polymerase is a thermostable DNA polymerase that allows multiple cycles of amplification without the need for new enzyme addition after each denaturation step.
How can the size of amplified DNA fragments be determined after PCR?
-Gel electrophoresis is used to visualize and estimate the size of amplified DNA fragments by comparing the gel bands to a molecular weight marker.
What are some factors that can interfere with a PCR reaction?
-Factors that can interfere with PCR include nucleotide composition of the DNA template, choice of DNA polymerase enzyme, buffer components, primer design, additives, and inhibitors.
How can PCR be optimized for different types of DNA templates?
-Optimizing PCR for different templates may involve adjusting the annealing temperature, using additives for GC-rich or AT-rich templates, and selecting the appropriate DNA polymerase and buffer.
What is the significance of depurination in long template amplification?
-Depurination, where the β-N-glycosidic bond in purine nucleoside is cleaved, can lead to incomplete replication as Taq DNA polymerase will not extend through apurinic positions, making it a limiting factor for long template amplification.
How can PCR inhibitors be managed in a reaction?
-PCR inhibitors can be managed by using sample-specific nucleic acid isolation protocols or by using additives that help reduce inhibition.
What resources does Applied Biological Materials (abm) offer for PCR?
-Applied Biological Materials offers a range of DNA polymerases, formulated PCR MasterMixes for each polymerase, and kits for easy DNA amplification from various samples such as plants, tissue, and blood.
Outlines

Esta sección está disponible solo para usuarios con suscripción. Por favor, mejora tu plan para acceder a esta parte.
Mejorar ahoraMindmap

Esta sección está disponible solo para usuarios con suscripción. Por favor, mejora tu plan para acceder a esta parte.
Mejorar ahoraKeywords

Esta sección está disponible solo para usuarios con suscripción. Por favor, mejora tu plan para acceder a esta parte.
Mejorar ahoraHighlights

Esta sección está disponible solo para usuarios con suscripción. Por favor, mejora tu plan para acceder a esta parte.
Mejorar ahoraTranscripts

Esta sección está disponible solo para usuarios con suscripción. Por favor, mejora tu plan para acceder a esta parte.
Mejorar ahoraVer Más Videos Relacionados

How we test for SARS-CoV-2 - RT-PCR (Reverse Transcription PCR)

Metode PCR (Polymerase Chain Reaction)

Pemeriksaan PCR (Polymerase Chain Reaction): Mekanisme dan Prinsip Dasar Pemeriksaan
Polymerase chain reaction (PCR) | Biomolecules | MCAT | Khan Academy

What is PCR? Polymerase Chain Reaction | miniPCR bio™

Prinsip dan Teknik PCR
5.0 / 5 (0 votes)
