DNA Sequencing Techniques | An Overview
Summary
TLDRThis lecture offers an in-depth overview of DNA sequencing techniques, explaining the process of determining the order of base pairs in DNA molecules. It covers the significance of DNA sequencing in detecting mutations, distinguishing organisms, and identifying human haplotypes. The video explores various sequencing methods, including direct sequencing techniques like Sanger sequencing and chemical sequencing, as well as next-generation sequencing technologies such as ion conductance, reversible dye terminator, sequencing by ligation, and nanopore sequencing. Each method is briefly explained, highlighting its unique approach to DNA analysis.
Takeaways
- 🧬 DNA sequencing is the process of determining the order of base pairs in a DNA molecule, crucial for understanding genetic information.
- 🔎 There are four bases in DNA: adenine (A), cytosine (C), guanine (G), and thymine (T), which are the building blocks of genetic code.
- 🔍 DNA sequencing helps in detecting mutations, distinguishing organisms, and identifying human haplotypes and polymorphisms.
- 🧪 Direct sequencing methods like chemical sequencing (Maxim Gilbert) and Sanger sequencing allow for direct reading of DNA base sequences.
- 🔬 Chemical sequencing uses chemical agents to cleave DNA at specific base pairs and involves electrophoresis to visualize and read the sequence.
- 🌟 Sanger sequencing is a modification of the DNA replication process using dideoxynucleotides (ddNTPs) to terminate DNA strand growth at specific bases.
- 🔄 Pyrosequencing is an indirect method that detects DNA sequence by monitoring light generation as nucleotides are incorporated into a growing DNA strand.
- 🧵 Bisulfite DNA sequencing is used to detect methylated cytosines, which play a role in gene regulation and chromatin structure.
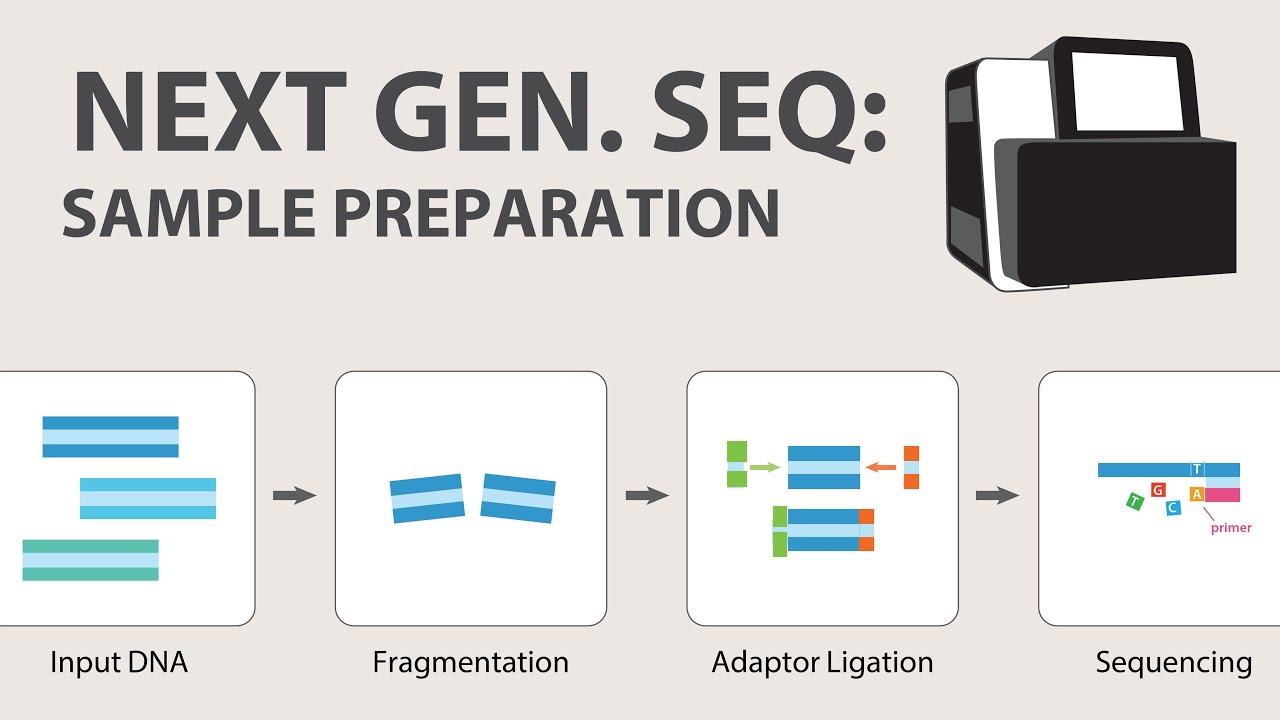
- 🌐 Next Generation Sequencing (NGS) technologies like ion conductance, reversible dye terminator, and nanopore sequencing allow for rapid, high-throughput DNA sequencing.
- 🧬 NGS techniques are used for large-scale genomic studies and require powerful computing for data analysis and assembly of sequenced libraries.
Q & A
What is DNA sequencing and why is it important?
-DNA sequencing is the process of determining the order of base pairs in a DNA molecule. It's important because it can help detect mutations in genes, distinguish between different organisms by analyzing specific gene sequences, and identify human haplotypes and polymorphisms, which provide insights into gene inheritance and function.
What are the four bases found in DNA and how are they abbreviated?
-The four bases found in DNA are adenine (A), cytosine (C), guanine (G), and thymine (T).
How does DNA sequencing help in identifying mutations?
-DNA sequencing helps identify mutations by determining the exact sequence of bases in a gene. A change in just one base can lead to a change in the gene's function, which can have significant effects on an individual.
What is the purpose of using the 16s rRNA gene in distinguishing microorganisms?
-The 16s rRNA gene is used to distinguish microorganisms because it contains universal sequences that are conserved across different species, allowing for the identification of specific differences that can differentiate one microorganism from another.
What are the two main types of direct DNA sequencing techniques mentioned in the script?
-The two main types of direct DNA sequencing techniques mentioned are chemical sequencing (Maxim Gilbert sequencing) and Sanger sequencing.
How does chemical sequencing differ from Sanger sequencing?
-Chemical sequencing uses chemical agents to cleave DNA at specific base pairs, while Sanger sequencing is a modification of the DNA replication process that uses modified dNTPs (ddNTPs) to create different lengths of DNA fragments.
What is the role of electrophoresis in DNA sequencing?
-Electrophoresis is used in DNA sequencing to separate DNA fragments based on their size. The separated fragments are then visualized and the sequence is read from the pattern they form on the gel or in the capillary.
Why is Sanger sequencing still widely used today?
-Sanger sequencing is still widely used because it can be adapted for automation, which allows for faster and more efficient sequencing. It uses fluorescent dyes instead of radioactive labels, making it safer and more convenient.
What is pyrosequencing and how does it determine the DNA sequence?
-Pyrosequencing is a DNA sequencing method that relies on the generation of light through luminescence whenever nucleotides are added to a growing strand of DNA. It determines the sequence by detecting which nucleotide generates a light signal when added.
What is bisulfite DNA sequencing and why is it used?
-Bisulfite DNA sequencing is used to detect methylated cytosines, which play a role in gene expression regulation and chromatin structure. It involves treating DNA with bisulfite to convert cytosines to uracil, leaving methylated cytosines unchanged, and then sequencing to identify the methylated sites.
What are the key features of Next Generation Sequencing (NGS) technologies?
-NGS technologies are characterized by their ability to sequence large numbers of templates carrying millions of bases in a short time. They use target libraries, require powerful computers and bioinformatics for data analysis, and include methods like ion conductance sequencing, reversible dye terminator sequencing, sequencing by ligation, and nanopore sequencing.
Outlines

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифMindmap

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифKeywords

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифHighlights

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифTranscripts

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тариф5.0 / 5 (0 votes)