Posttranscriptional Modification of RNA | RNA processing | Biochemistry
Summary
TLDRThis video discusses the post-transcriptional modification of RNA, comparing the processes in eukaryotes and prokaryotes. It explains how RNA processing occurs in eukaryotic cells, involving the addition of a 5' cap, a poly-A tail, and intron removal through splicing. The video also covers the differences in ribosomal RNA (rRNA) and transfer RNA (tRNA) processing, including enzyme involvement, base modifications, and how these modifications contribute to RNA stability. By highlighting key steps in RNA processing, the video provides a clear understanding of how genetic information is prepared for protein synthesis.
Takeaways
- 🧬 RNA processing differs between eukaryotes and prokaryotes, with eukaryotes requiring more extensive modifications.
- 📚 In eukaryotes, transcription occurs in the nucleus, while translation happens in the cytoplasm, unlike prokaryotes where both occur in the cytoplasm.
- 🌟 Eukaryotic mRNA undergoes co-transcriptional and post-transcriptional modifications, whereas prokaryotic mRNA does not require processing.
- 🔍 Both eukaryotic and prokaryotic rRNA and tRNA require post-transcriptional modifications.
- 🧪 Pre-ribosomal RNA in prokaryotes is synthesized by RNA polymerase holoenzyme, while in eukaryotes it's synthesized by RNA polymerase I.
- 📏 Eukaryotic 5S rRNA is synthesized separately by RNA polymerase III and undergoes independent modification.
- 🧬 Ribosomal RNA processing involves cleavage by ribonucleases and trimming by exonucleases to produce intermediate-sized RNA.
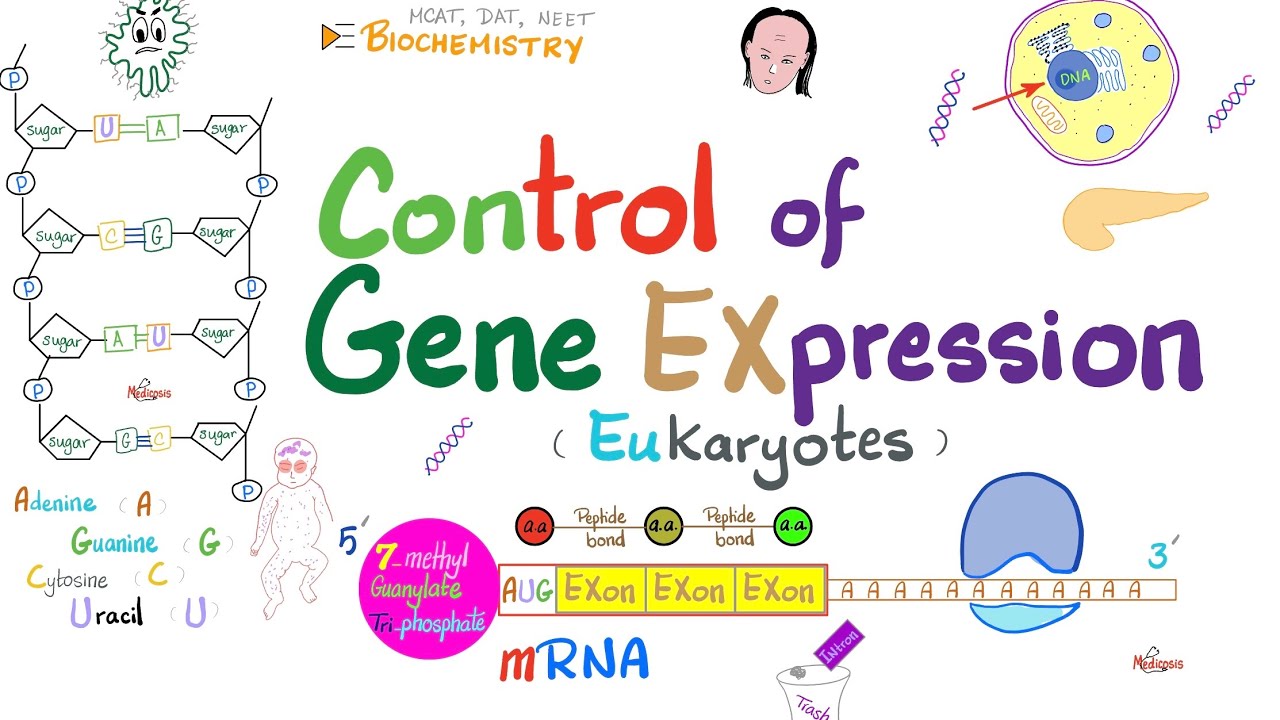
- 📝 mRNA processing in eukaryotes includes the addition of a 5' cap, a poly(A) tail, and the removal of introns.
- 🔑 The 5' cap protects mRNA from degradation and promotes binding to the ribosome, while the poly(A) tail stabilizes mRNA in the cytoplasm.
- ✂️ Splicing, the removal of introns and joining of exons, requires small nuclear RNAs and occurs in a specialized structure called the spliceosome.
- 🔄 tRNA processing involves cleavage by ribozyme RNA sp, modification of uracil residues, and the addition of a CCA sequence at the 3' end for amino acid attachment.
Q & A
What is RNA processing in eukaryotes?
-RNA processing in eukaryotes involves modifications to the primary RNA transcript, including co-transcriptional and post-transcriptional modifications, such as the addition of a 5' cap, polyadenylation, and removal of introns.
How does RNA processing differ between eukaryotes and prokaryotes?
-In eukaryotes, RNA processing occurs in the nucleus and involves modifications such as capping, polyadenylation, and splicing. In prokaryotes, RNA processing is not needed as transcription and translation occur simultaneously in the cytoplasm.
What enzymes are involved in synthesizing ribosomal RNA (rRNA) in eukaryotes and prokaryotes?
-In eukaryotes, ribosomal RNA is synthesized by RNA polymerase I, while in prokaryotes, it is synthesized by RNA polymerase holoenzyme.
What is the role of the 5' cap in mRNA processing?
-The 5' cap, a 7-methyl guanosine (m7G), is added to the 5' end of mRNA and protects it from degradation, as well as promotes ribosome binding for translation.
What is the significance of the poly-A tail in mRNA processing?
-The poly-A tail, added to the 3' end of mRNA, provides stability to the mRNA in the cytoplasm and helps in efficient export from the nucleus and translation.
How are introns removed from the pre-mRNA in eukaryotes?
-Introns are removed from pre-mRNA by the splicing process, which involves small nuclear RNAs (snRNAs) that form complexes with proteins to create spliceosomes, which then remove the introns.
What is the function of small nuclear ribonucleoproteins (snRNPs) in splicing?
-snRNPs, such as U1, U2, U4, U5, and U6, recognize specific sequences at the intron-exon boundaries and catalyze the splicing of pre-mRNA by forming the spliceosome complex.
What sequence motifs are found at the 5' and 3' ends of introns?
-The 5' end of introns has a GU sequence, while the 3' end has an AG sequence, which are recognized by snRNPs during splicing.
What role does the branch site play in the splicing process?
-The branch site contains an adenosine residue with a 2' hydroxyl group, which initiates a nucleophilic attack on the 5' splice site, forming a lariat structure during splicing.
How is pre-transfer RNA (tRNA) processed in eukaryotes?
-Pre-tRNA is processed by removing a 16-nucleotide sequence from the 5' end, replacing the 3' uracil residues with a CCA sequence, and removing a 14-nucleotide intron from the anticodon loop.
Outlines

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantMindmap

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantKeywords

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantHighlights

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantTranscripts

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenant5.0 / 5 (0 votes)