Biotechniques | Principles of Primer Design for Full Gene Amplification
Summary
TLDRIn this Catalyst University video, Kevin discusses primer design for PCR, focusing on amplifying the entire gene sequence of an actin protein. He explains the importance of identifying start and stop codons and demonstrates how to use online tools like Primer-BLAST for primer optimization. However, for full gene amplification, he advises manually selecting primers based on the 5' and 3' ends and using a reverse complement tool to ensure the entire gene sequence is captured for applications like cloning.
Takeaways
- 🔬 Primer design is crucial for PCR to ensure specific gene amplification.
- 🧬 The presence of thymine (T) indicates the sequence is DNA, not RNA.
- 📖 The start codon (ATG) and stop codon (TGA) in mRNA signify the boundaries of a gene.
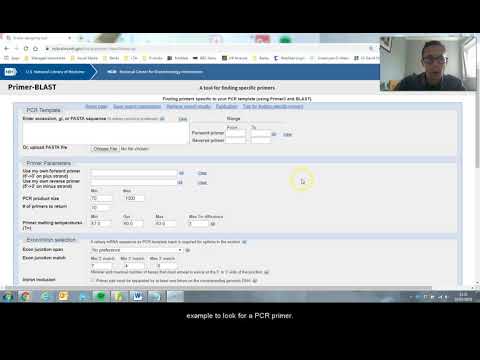
- 🔍 Primer-BLAST is a useful online tool for optimizing primer design.
- 📏 Typically, primers should be around 20 nucleotides long for effective PCR.
- 🔄 To amplify the entire gene, the forward primer should start from the initial methionine coding nucleotide triplet.
- 🔄 The reverse primer sequence is obtained by finding the reverse complement of a selected sequence from the 3' end.
- 🌐 Websites like primer-blast and Harvard's get reverse complement tool facilitate primer design.
- 🔄 Primer-BLAST is more suitable for partial gene sequences, not full gene amplification.
- 🔬 For full gene amplification, manually designing primers based on the 5' and 3' ends of the gene is necessary.
- 📝 Primer sequences can vary in length, with some experiments using primers up to 40 nucleotides.
Q & A
What is the main topic of the video?
-The main topic of the video is the principles of primer design for PCR (Polymerase Chain Reactions), specifically focusing on how to obtain specific forward and reverse primers from a given DNA sequence.
Why does the presence of thymine (T) indicate that the sequence is DNA?
-Thymine (T) is unique to DNA, whereas RNA contains uracil (U) instead. The presence of T's in the sequence indicates that it is a DNA sequence.
What does the ATG codon represent in the context of the video?
-In the video, the ATG codon represents the start codon for the gene sequence, which codes for methionine in the protein synthesis process.
What is the purpose of using a primer design website like Primer-BLAST?
-Primer-BLAST is used to optimize the design of primers for PCR reactions by providing parameters such as minimum and maximum melting temperatures and product size.
Why might the primer pairs suggested by Primer-BLAST not be suitable for amplifying the entire gene?
-The primer pairs suggested by Primer-BLAST might not amplify the entire gene because the optimization process focuses on specific parameters that may not align with the goal of obtaining the full gene sequence.
How does the video suggest obtaining the forward primer sequence?
-The video suggests obtaining the forward primer sequence by starting from the first nucleotide of the coding strand and counting over approximately 20 nucleotides.
What is the significance of the reverse complement of a DNA sequence in primer design?
-The reverse complement of a DNA sequence is significant in primer design because it allows for the creation of the reverse primer, which is necessary to amplify the entire gene sequence from the 3' end.
How does the video demonstrate obtaining the reverse primer sequence?
-The video demonstrates obtaining the reverse primer sequence by copying a 20-nucleotide sequence from the 3' end of the gene, pasting it into a website to find the reverse complement, and then using that sequence as the reverse primer.
What are the typical lengths of primers used in PCR reactions?
-The video mentions that typical primer lengths are around 20 nucleotides, but they can vary, with some primers being as long as 25, 30, or even 40 nucleotides.
What are some applications of amplifying the entire gene sequence as discussed in the video?
-Some applications of amplifying the entire gene sequence include cloning the gene, forcing its expression in a bacterium, and using it for restriction digests.
How does the video recommend finding the reverse complement of a DNA sequence?
-The video recommends using a specific website or searching for 'get reverse complement' on Google, which leads to a Harvard website that provides this service.
Outlines

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantMindmap

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantKeywords

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantHighlights

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenantTranscripts

Cette section est réservée aux utilisateurs payants. Améliorez votre compte pour accéder à cette section.
Améliorer maintenant5.0 / 5 (0 votes)