Sekuensing DNA Menggunakan Aplikasi MEGA 11
Summary
TLDRThis tutorial demonstrates the use of MEGA (Molecular Evolutionary Genetics Analysis) software for DNA sequence analysis and phylogenetic tree construction. The speaker guides viewers through the process of uploading, trimming, and aligning DNA sequences, followed by using an online tool to identify species. They then explain how to create a phylogenetic tree to visualize evolutionary relationships between species based on their genetic data. The tutorial emphasizes the importance of understanding genetic sequence relationships and provides step-by-step instructions for handling sequence data and interpreting phylogenetic results.
Takeaways
- 😀 The speaker introduces themselves as part of group 3 in a biotechnology practicum and begins discussing the Mega 11 (molecular evolutionary analysis) application.
- 😀 The process starts by selecting 'Edit or build' and choosing 'Create new Element' to begin building a new DNA sequence.
- 😀 The speaker explains how to upload DNA sequence data and clean up chromatograms by trimming poor-quality data from both ends of the sequence.
- 😀 After trimming the sequence, the speaker saves it and exports it in FASTA format for further analysis.
- 😀 The speaker uses a website (CDI) to upload the trimmed DNA sequence to identify the species based on lobster data, ensuring the correct organism is selected.
- 😀 The alignment of the DNA sequence is carried out by renaming species and performing a sequence alignment with a matrix to identify matching sequences.
- 😀 The speaker explains how to interpret the results, highlighting the presence of stars in the alignment matrix, which indicates matching sequences across species.
- 😀 The next step involves constructing a phylogenetic tree using Mega software by selecting the appropriate method and setting the parameters for analysis.
- 😀 The phylogenetic tree is generated, and the speaker shows how to label and color-code the lobster sequence within the tree for clarity.
- 😀 The speaker explains how to interpret the phylogenetic tree, noting that closer species are more related, and further species have more distant evolutionary ties.
- 😀 The final steps involve saving the phylogenetic tree as a PNG file, which can be used for further study or presentation purposes.
Q & A
What is the main objective of the tutorial described in the script?
-The main objective is to demonstrate how to use the Mega 11 software for molecular evolutionary analysis, including DNA sequence editing, alignment, and phylogenetic tree construction.
How does the user begin using the Mega 11 software?
-The user starts by selecting the 'Edit or Build' option and creating a new DNA element within the software to begin their analysis.
What does the 'Mask' function do in Mega 11?
-The 'Mask' function is used to trim or cut portions of the chromatogram that are of lower quality, ensuring that only the high-quality sections are analyzed.
Why does the user trim the DNA sequence in the tutorial?
-The user trims the DNA sequence to remove low-quality areas at both ends of the chromatogram, focusing only on the high-quality portion for further analysis.
What is the purpose of uploading the sequence to the Lobster 2.1 website?
-Uploading the sequence to the Lobster 2.1 website allows the user to align the DNA sequences and identify the species based on the molecular data.
What does a star (*) symbol indicate in the alignment results?
-The star (*) symbol indicates that the sequence is conserved across all species being compared, highlighting regions of similarity in the DNA sequences.
What kind of analysis does the Mega 11 software perform to construct a phylogenetic tree?
-Mega 11 constructs the phylogenetic tree using a substitution model (e.g., Transition plus Transversion) to assess evolutionary relationships between the sequences.
How does the user interpret the phylogenetic tree once it is generated?
-The user interprets the phylogenetic tree by analyzing the proximity of the branches: species with closer branches are more closely related, while distant branches indicate a more distant evolutionary relationship.
What is the significance of marking species in the phylogenetic tree with different colors?
-Marking species with different colors or shapes helps to visually distinguish them in the phylogenetic tree, making it easier to interpret the relationships between different species.
How does the user save the final phylogenetic tree after creating it?
-The user saves the phylogenetic tree by selecting the 'Image' option, saving it as a PNG file, and naming the file (e.g., 'Phylogenetic Tree') before storing it in a desired location.
Outlines

此内容仅限付费用户访问。 请升级后访问。
立即升级Mindmap

此内容仅限付费用户访问。 请升级后访问。
立即升级Keywords

此内容仅限付费用户访问。 请升级后访问。
立即升级Highlights

此内容仅限付费用户访问。 请升级后访问。
立即升级Transcripts

此内容仅限付费用户访问。 请升级后访问。
立即升级浏览更多相关视频
Tutorial BioEdit: konsensus sekuen DNA hasil sekuensing dengan bioedit. Lengkap dan pasti bisa!!!

ANALISIS FENETIK DAN FILOGENETIK

PYTHON Programming in Bioinformatics| Bioinformatics Research Projects Using Python for Freshers
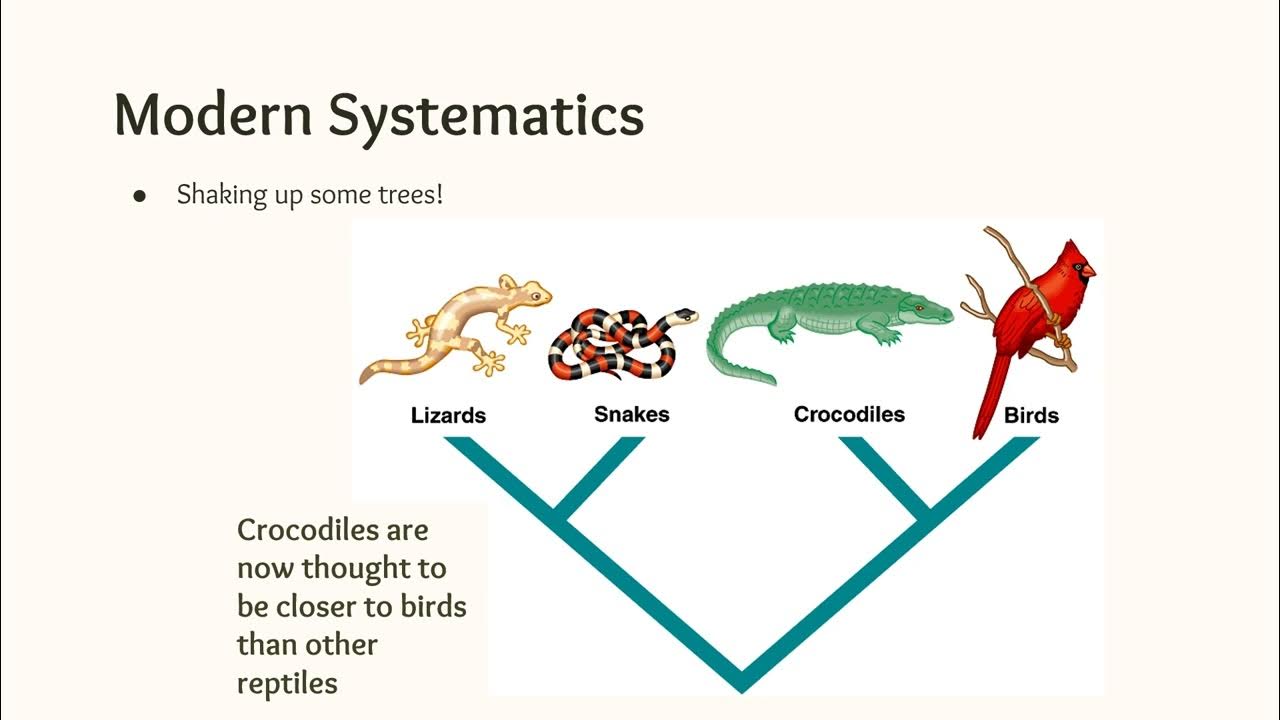
Phylogeny and Cladistics

NCBI Minute: Five Teaching Examples Using BLAST
Training Trees
5.0 / 5 (0 votes)
