RNA Seq: Principle and Workflow of RNA Sequencing
Summary
TLDRThis video delves into RNA-Seq, a next-generation sequencing technique that uncovers RNA presence and quantity in biological samples. It traces RNA-Seq's evolution from Sanger sequencing to Illumina's dominance and emerging third-generation technologies. RNA-Seq offers high-throughput, single-base resolution profiling, overcoming microarray and Sanger sequencing limitations. The video outlines the workflow, from RNA isolation to data generation, and touches on challenges like PCR bias. It also discusses the importance of quality assessment and the technology's ability to detect sequence variations and gene regulation.
Takeaways
- 🧬 **RNA-Seq Definition**: RNA sequencing (RNA-Seq) uses next-generation sequencing (NGS) to analyze the presence and quantity of RNA in a sample at a specific developmental stage or physiological condition.
- 📈 **Development of RNA-Seq**: RNA-Seq evolved from Sanger sequencing and microarrays to become dominant with the advent of NGS technologies like Illumina and Roche 454.
- 🔎 **Advantages of RNA-Seq**: RNA-Seq offers high-throughput RNA profiling at single base resolution with low background noise, surpassing microarrays and Sanger sequencing in sensitivity and specificity.
- 🧪 **Third-Generation Sequencing**: Third-generation sequencing technologies like Pacific Biosciences provide long-read sequencing, useful for identifying new transcripts and isoforms without fragmentation.
- 📉 **Challenges in RNA-Seq**: Challenges include short read biases and PCR amplification biases that can affect gene expression quantitation.
- 🧪 **Workflow of RNA-Seq**: The workflow involves converting RNA into cDNA fragments, attaching sequencing adapters, generating sequence data, and aligning reads to a reference genome or transcriptome.
- 🌟 **Types of Reads**: RNA-Seq classifies reads into exonic, junction, and poly-A reads to generate a base resolution expression profile.
- 🧪 **Sample Collection**: Total RNA is isolated and processed, often involving ribosomal RNA depletion to enrich non-ribosomal RNA species.
- 🔬 **Fragmentation**: RNA molecules are fragmented into smaller pieces for sequencing, with cDNA fragmentation providing information about the 3' ends and RNA fragmentation accessing the transcript body.
- 📊 **Bioinformatics Analysis**: The first step in RNA-Seq analysis is quality assessment, followed by read mapping or assembly, and gene expression level inference.
- 🔍 **Applications**: RNA-Seq is used to detect differential expression, SNPs, fusion genes, and post-transcriptional gene regulation.
Q & A
What is RNA-seq and what does it reveal about biological samples?
-RNA-seq, short for RNA sequencing, uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a specific developmental stage or physiological condition. It is a powerful tool for analyzing the dynamic cellular transcriptome, which is essential for interpreting the functional elements of the genome, revealing molecular constituents of cells and tissues, and understanding development and disease.
What technological advancements have influenced the development of RNA-seq?
-The development of RNA-seq has been influenced by several technological advancements, including Sanger sequencing technology, microarrays, and the advent of next-generation sequencing technologies. The first RNA-seq paper was published in 2006 using Roche 454 technology, and the technology matured with the advent of Illumina technology in 2008. Third-generation sequencing technologies, such as Pacific Biosciences' long-read sequencing, are also emerging in RNA-seq.
How does third-generation sequencing technology impact RNA-seq?
-Third-generation sequencing technology, such as Pacific Biosciences' long-read sequencing, allows for full-length transcript sequencing without the need for fragmentation. This is useful for identifying new transcripts and novel isoforms, alternative splicing sites, fusion gene expression, and allelic expression more accurately.
What are the advantages of RNA-seq over traditional microarray and Sanger sequencing?
-RNA-seq has several advantages over traditional microarray and Sanger sequencing, including high throughput RNA profiling at single base resolution, low background noise, the ability to map transcribed regions and gene expression simultaneously, and the capability to distinguish different isoforms and allelic expression. It also requires a lower amount of RNA and is less costly.
What are the challenges faced by RNA-seq?
-RNA-seq faces challenges such as short read biases and PCR biases. Short read lengths were a concern, but Illumina sequencing technology has increased read length and throughput. PCR amplification can impact the accuracy of gene expression quantitation, but amplification-free technologies and PCR-free methods for Illumina sequencing have been developed to address this.
What is the workflow of RNA-seq using high-throughput sequencing technology?
-The workflow of RNA-seq involves converting long RNAs into a library of cDNA fragments through RNA or DNA fragmentation, attaching sequencing adapters to each cDNA fragment, and generating sequence data from both ends in a high-throughput manner. The resulting sequence reads are aligned with the reference genome or transcriptome and classified into exonic, junction, and poly-A reads to generate a base resolution expression profile.
How is total RNA isolated for RNA-seq?
-Total RNA is usually isolated via organic extraction or silica membranes of spin columns. The total RNA sample is then processed either by direct selection of polyA RNA or by selective removal of ribosomal RNA, as ribosomal RNA is often not the research focus and can greatly reduce the coverage of useful transcripts.
Why is ribosomal RNA depletion preferred over polyA RNA selection?
-Ribosomal RNA depletion is preferred over polyA RNA selection because it enriches all non-ribosomal RNA species, including tRNA, ncRNA, non-polyA mRNA, and pre-processed RNA. This approach captures a broader range of RNA species compared to polyA RNA selection, which may miss some RNA transcripts that lack polyA tails.
How are larger RNA molecules prepared for deep sequencing technologies?
-Larger RNA molecules need to be fragmented into smaller pieces (200 to 500 nt) before deep sequencing technologies. cDNA fragmentation is biased towards the identification of sequences from the 3' ends of transcripts, while RNA fragmentation provides access to the precise identity of the transcript body.
What are the different platforms that dominate RNA-seq?
-RNA-seq is currently dominated by three different platforms: Illumina, Ion Torrent (Thermo Fisher Scientific), and Pacific Biosciences' SMRT. These platforms offer varying read lengths and technologies for sequencing.
How does RNA-seq enable the detection of differential expression and other genomic features?
-RNA-seq allows for the detection of differential expression across treatments or conditions by normalizing the differences between samples. It also enables the identification of SNPs, fusion genes, and post-transcriptional gene regulation, such as RNA editing, degradation, and translation.
Outlines

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифMindmap

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифKeywords

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифHighlights

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифTranscripts

Этот раздел доступен только подписчикам платных тарифов. Пожалуйста, перейдите на платный тариф для доступа.
Перейти на платный тарифПосмотреть больше похожих видео
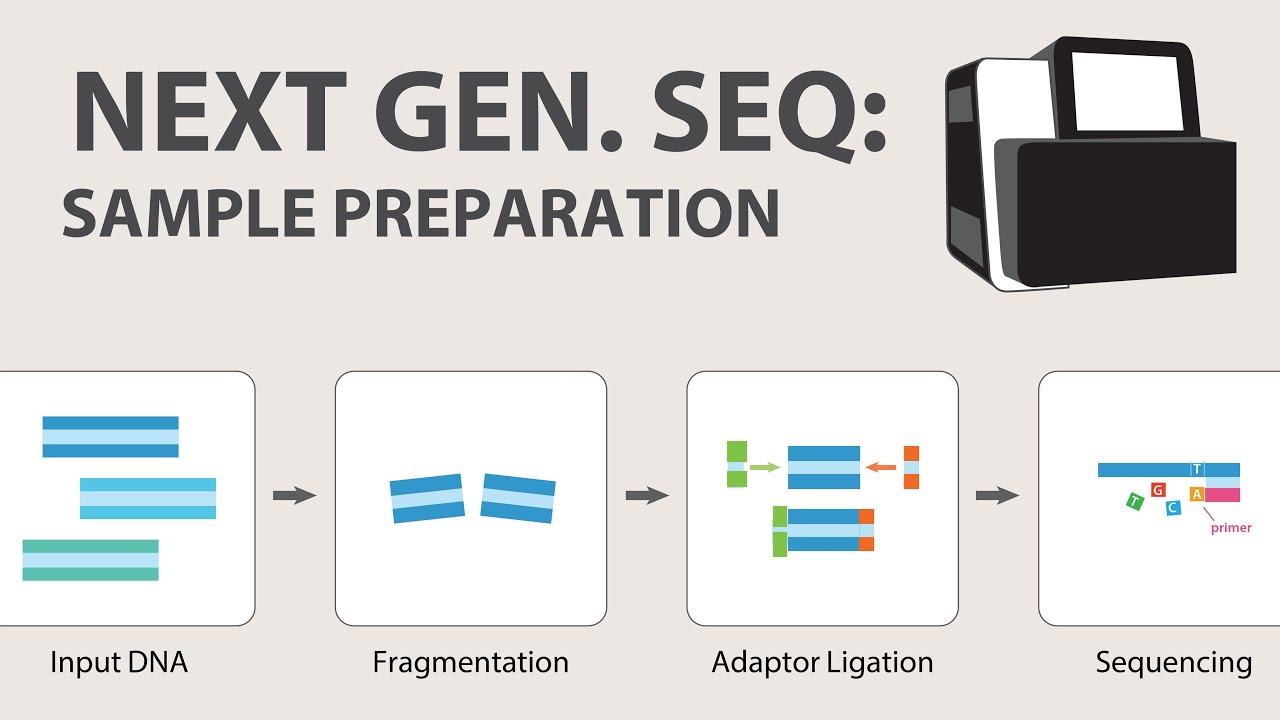
2) Next Generation Sequencing (NGS) - Sample Preparation

Intro to Proteomics / Mass Spectrometry (MS)

Transcription in eukaryotes | Chromatin-centric view of transcription | RNA pol II transcripts

12 Quick Tips for NGS Library Preparation
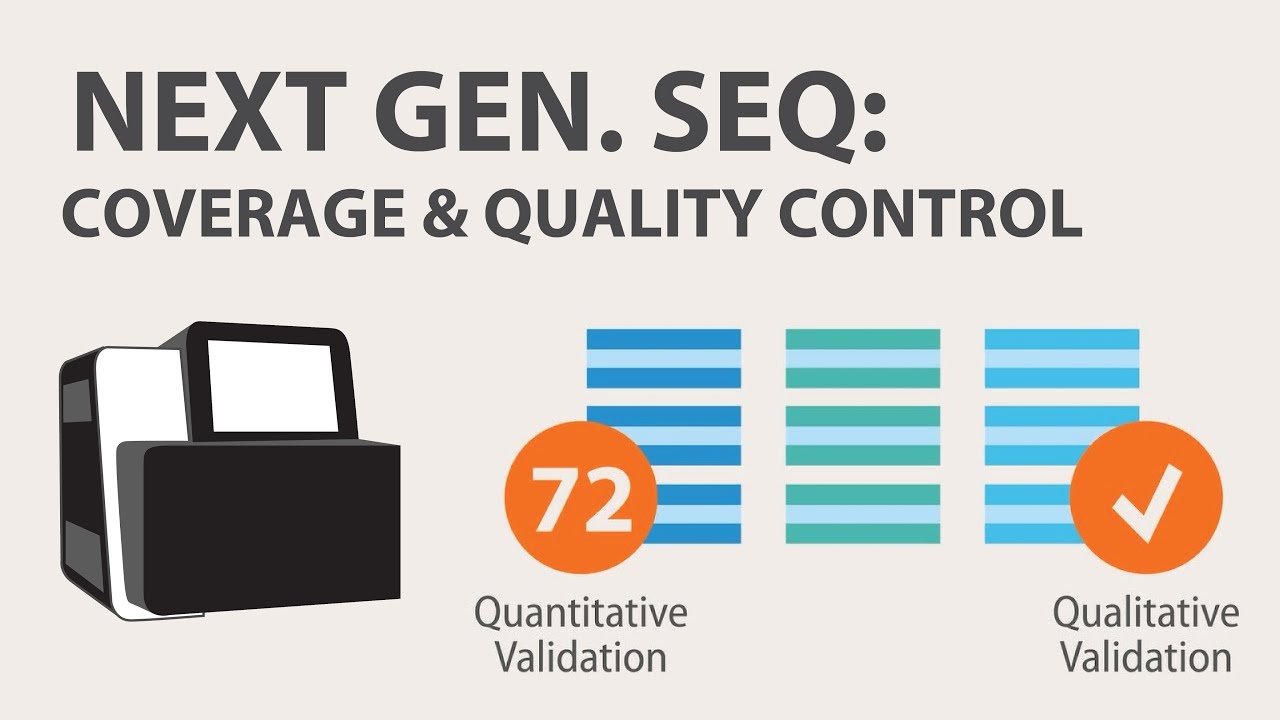
3) Next Generation Sequencing (NGS) - Coverage & Sample Quality Control

RNA sequencing
5.0 / 5 (0 votes)
