Intro to Proteomics / Mass Spectrometry (MS)
Summary
TLDRThis video provides an overview of proteomics, explaining how it differs from methods like RNA-seq and Drop-seq. It walks through the central dogma of biology, focusing on the transition from DNA to RNA to protein, and highlights the complexities of protein structures, including post-translational modifications. The video then covers the experimental workflow for proteomic studies, emphasizing the use of mass spectrometry to analyze proteins in biological samples. The process is similar to RNA-seq but focuses on protein identification and quantification. It also touches on advanced techniques such as protein interaction studies and single-cell proteomics.
Takeaways
- 😀 Proteomics is a high-throughput method used to measure all proteins in a cell or tissue sample, providing a broader view of cellular activity than RNA-seq.
- 😀 The central dogma of biology involves DNA being transcribed into RNA, which is then translated into proteins, but mRNA is not always an accurate reflection of protein expression.
- 😀 Proteins undergo several stages of folding, from primary structure (a polypeptide chain) to secondary (alpha helices and beta sheets), tertiary, and quaternary structures that define their function.
- 😀 Post-translational modifications, such as phosphorylation, can alter protein function, activating or inactivating proteins depending on the modification.
- 😀 Unlike RNA-seq, which measures mRNA, proteomics directly measures protein abundance and provides more complex insights into protein levels and interactions.
- 😀 Proteomics is often used in combination with mass spectrometry (MS) to identify and quantify proteins, where the mass-to-charge ratio helps determine protein presence and abundance.
- 😀 Data from mass spectrometry are processed into count tables, and statistical analysis (e.g., Wilcoxon test, t-test) helps identify significant changes in protein levels between experimental conditions.
- 😀 Visualizations like heatmaps and volcano plots are used to interpret proteomics data and identify proteins of interest based on statistical significance.
- 😀 A significant difference between RNA-seq and proteomics is that RNA-seq measures gene expression, while proteomics quantifies proteins and their interactions in the cell.
- 😀 Immunoprecipitation (IP) is used to study protein-protein interactions by isolating specific proteins and analyzing their binding partners, providing insight into protein functions within cells.
- 😀 The emerging field of single-cell proteomics allows for the analysis of protein expression at the single-cell level, offering deeper insights into cellular heterogeneity and protein function in individual cells.
Q & A
What is the central dogma of biology, and how does it relate to proteomics?
-The central dogma of biology describes the flow of genetic information: DNA is transcribed into RNA, and RNA is translated into protein. In proteomics, we focus on the protein level, which is the final product of the central dogma and represents the active molecules within the cell.
Why is proteomics considered more complex than RNA-seq?
-Proteomics is more complex than RNA-seq because proteins can undergo multiple post-translational modifications and fold into different structures, resulting in a far greater diversity of forms than the transcriptome. The complexity increases further as proteins interact in intricate networks, which RNA-seq does not capture.
What are the different levels of protein structure, and why are they important in proteomics?
-Proteins have four structural levels: primary (amino acid chain), secondary (alpha helices and beta sheets), tertiary (3D folding), and quaternary (multiple chains forming a functional unit). These structures determine the protein's function, and proteomics aims to understand these details to elucidate cellular processes.
What role do post-translational modifications play in protein function?
-Post-translational modifications, such as phosphorylation, acetylation, or glycosylation, are chemical changes to proteins after they are synthesized. These modifications can activate or deactivate proteins, influence their stability, or alter their interactions with other molecules, thus playing a crucial role in regulating cellular activities.
How does mass spectrometry work in proteomics?
-Mass spectrometry works by ionizing protein samples and measuring the deflection of ions in a magnetic field. The deflection is inversely proportional to their mass-to-charge ratio, allowing for the identification and quantification of proteins based on their unique mass spectrum profiles.
What is the significance of a mass spectrum in proteomics?
-A mass spectrum provides a visual representation of protein ions based on their mass-to-charge ratio. The height of the peaks represents the abundance of specific proteins, and the number of peaks indicates the different protein isoforms or variants present in the sample.
How do researchers analyze proteomics data after mass spectrometry?
-After obtaining mass spectrometry data, researchers analyze it by generating a count table that lists protein identities and their relative abundance. Statistical algorithms, like Wilcoxon or T-test, are then used to identify significant changes in protein expression, and visualizations such as heat maps and volcano plots are created to help interpret the data.
What is the importance of controls in proteomics experiments?
-Controls are crucial in proteomics experiments to ensure that any observed differences in protein expression are due to the experimental condition rather than external variables. A well-chosen control helps validate the results and ensures the reliability of the findings.
What is the difference between proteomics and RNA-seq in terms of methodology and output?
-Proteomics uses mass spectrometry to measure proteins, providing both qualitative (which proteins are present) and quantitative (how much of each protein is present) data. In contrast, RNA-seq measures RNA levels, offering insights into gene expression but not directly into the functional proteins produced by those genes.
What is the concept of protein-protein interaction studies in proteomics, and how are they conducted?
-Protein-protein interaction studies in proteomics involve isolating a protein of interest (e.g., using an antibody) and identifying other proteins that bind to it. This allows researchers to explore cellular networks and understand how proteins work together. Techniques like immunoprecipitation (IP) combined with mass spectrometry are commonly used for this purpose.
How is single-cell proteomics different from traditional proteomics?
-Single-cell proteomics allows researchers to measure protein levels in individual cells, offering a higher resolution of data. This technique can reveal cellular heterogeneity and provide insights into protein expression at a much finer scale than traditional bulk proteomics, where protein levels are averaged across a population of cells.
Outlines

このセクションは有料ユーザー限定です。 アクセスするには、アップグレードをお願いします。
今すぐアップグレードMindmap

このセクションは有料ユーザー限定です。 アクセスするには、アップグレードをお願いします。
今すぐアップグレードKeywords

このセクションは有料ユーザー限定です。 アクセスするには、アップグレードをお願いします。
今すぐアップグレードHighlights

このセクションは有料ユーザー限定です。 アクセスするには、アップグレードをお願いします。
今すぐアップグレードTranscripts

このセクションは有料ユーザー限定です。 アクセスするには、アップグレードをお願いします。
今すぐアップグレード関連動画をさらに表示
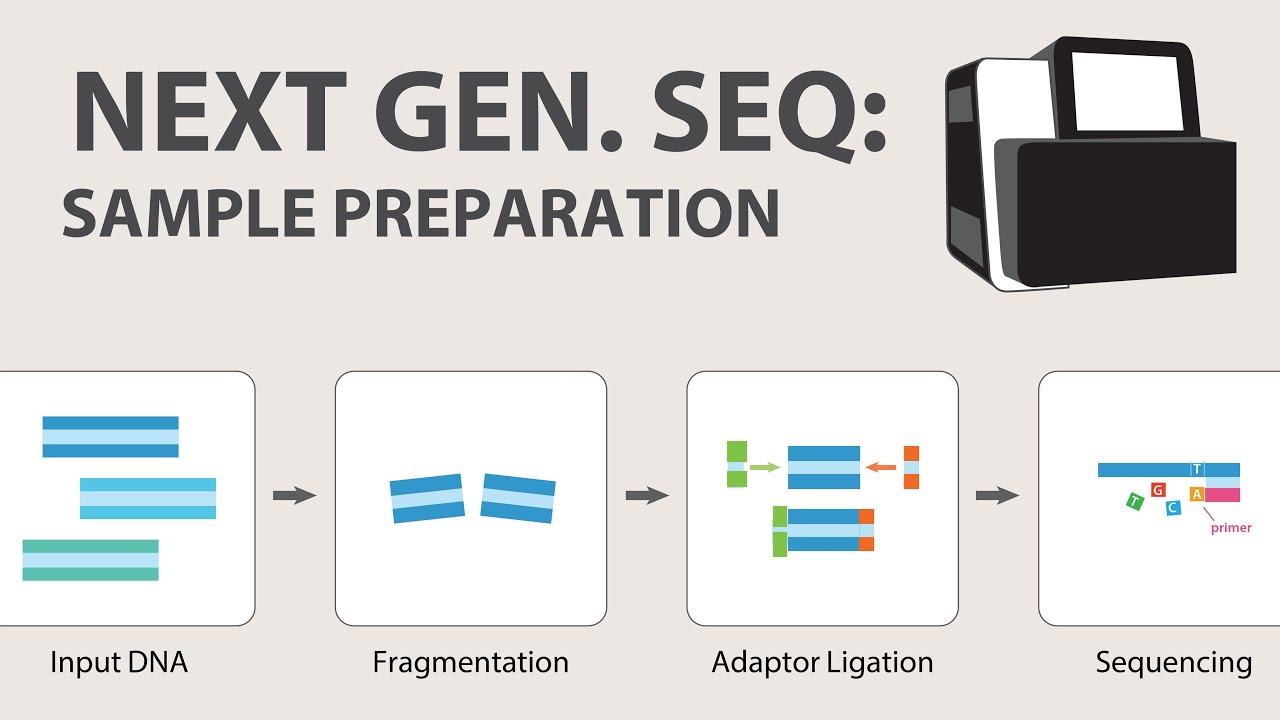
2) Next Generation Sequencing (NGS) - Sample Preparation

RNA Seq: Principle and Workflow of RNA Sequencing

The Beginner's Guide to RNA-Seq - #ResearchersAtWork Webinar Series

Epigenetics3: Histone Modification and ChIP-seq

Transcription in eukaryotes | Chromatin-centric view of transcription | RNA pol II transcripts

Dna Replication Part 2
5.0 / 5 (0 votes)
