RNA Transcription 3b'
Summary
TLDRThis lecture delves into the intricacies of transcription and RNA processing, focusing on the initiation phase and the pivotal role of sigma factors in recognizing promoter sequences. It discusses the transition from initiation to elongation, akin to DNA replication, where RNA polymerase reads the template strand and synthesizes RNA. The session also covers the mechanisms of transcription termination in prokaryotes, contrasting intrinsic and rRNA-dependent termination. The lecture further explores the complexities of eukaryotic transcription, highlighting the involvement of multiple RNA polymerases, general transcription factors, and the role of enhancers and silencers in regulating the process.
Takeaways
- 🔬 The lecture discusses the process of transcription and RNA processing, focusing on the initiation, elongation, and termination of transcription in both prokaryotes and eukaryotes.
- 🌟 The importance of the sigma factor in prokaryotes is highlighted, as it recognizes promoter sequences and initiates transcription.
- 🧬 Elongation in transcription is compared to DNA replication, with RNA polymerase moving along the template strand and synthesizing RNA in a 5' to 3' direction.
- 🔄 Termination of transcription in prokaryotes can occur through two mechanisms: intrinsic (row-independent) and extrinsic (row-dependent), involving specific sequences and proteins.
- 🧲 Intrinsic termination involves a hairpin loop and a string of A's on the template strand, which causes RNA polymerase to pause and release the RNA.
- 🔄 Extrinsic termination in prokaryotes relies on the rho protein, which binds to the RNA at a specific site and disrupts the DNA-RNA hybrid to terminate transcription.
- 🌐 Eukaryotic transcription is more complex, involving a larger genome, the presence of a nucleus, and the need for RNA processing and export to the cytoplasm.
- 🧬 Eukaryotes have three different RNA polymerases, each with distinct promoter sequences and functions, including the transcription of ribosomal genes, small RNA genes, and protein-coding genes.
- 🔑 Transcription factors play a crucial role in eukaryotic transcription, binding to promoter elements and facilitating the binding of RNA polymerase to initiate transcription.
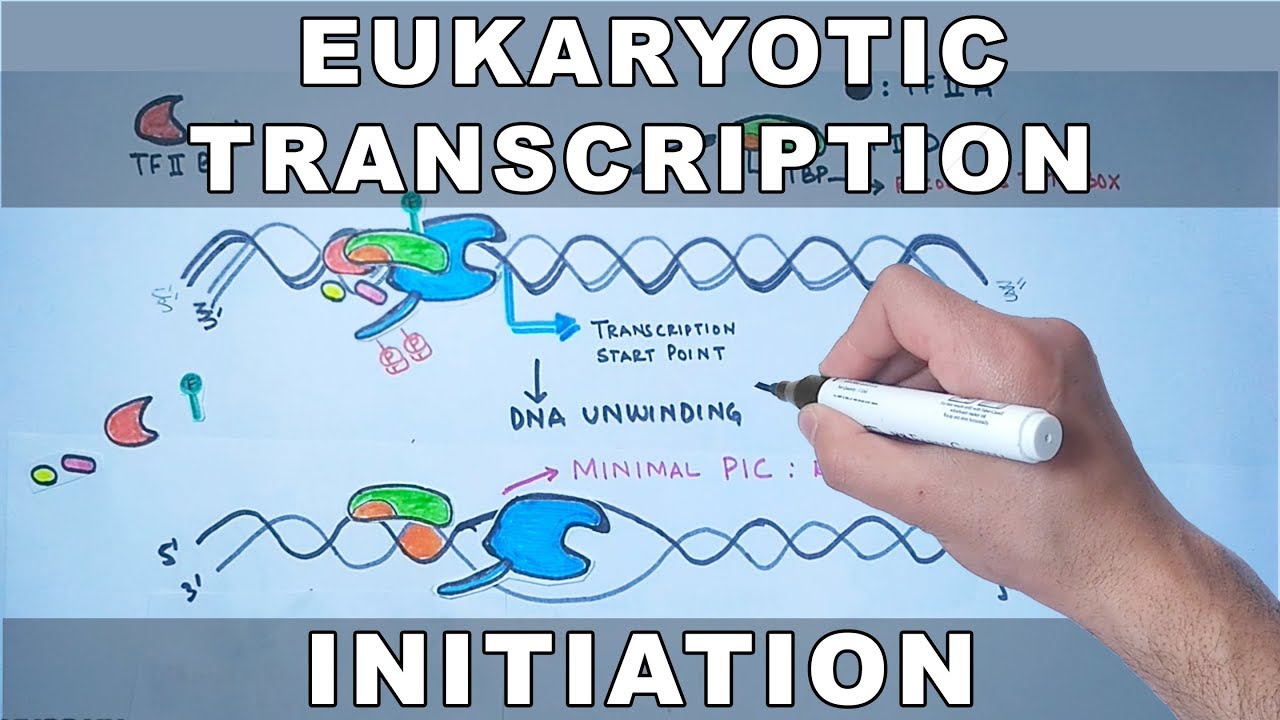
- 🔋 The process of transcription initiation in eukaryotes involves a specific order of transcription factor binding, leading to the formation of a pre-initiation complex and the start of transcription.
Q & A
What is the role of the sigma factor in the initiation of transcription?
-The sigma factor is crucial for the initiation of transcription as it helps the RNA polymerase recognize and bind to the promoter sequences, specifically the -10 (Pribnow box) and -35 regions, enabling the start of RNA synthesis.
How does the elongation process of transcription differ between DNA replication and RNA synthesis?
-Although elongation in transcription is similar to DNA replication, the key difference is that RNA polymerase moves along the DNA template strand from 3' to 5', synthesizing RNA in the 5' to 3' direction using ribonucleotide triphosphates instead of deoxyribonucleotides.
What is the function of the 'transcription bubble' mentioned in the script?
-The 'transcription bubble' is a region of local denaturation that forms behind the RNA polymerase as it moves along the DNA template. It consists of the single-stranded DNA and the newly synthesized RNA, and it allows for the continuous synthesis of RNA while the DNA re-anneals behind the bubble.
What are the two mechanisms of transcription termination in prokaryotes?
-The two mechanisms of transcription termination in prokaryotes are Rho-independent termination and Rho-dependent termination. Rho-independent termination involves the formation of a hairpin loop and a string of U's in the RNA, causing the RNA polymerase to pause and release the RNA. Rho-dependent termination requires the Rho protein, which binds to a specific site on the RNA and moves along it until it reaches the RNA polymerase, causing termination.
What is the significance of the poly U stretch in RNA during Rho-independent termination?
-The poly U stretch in RNA, resulting from a string of A's on the template strand, plays a role in Rho-independent termination by creating weak A-U base pairing that facilitates the release of the RNA from the DNA template, thus ending transcription.
How does the Rho protein contribute to Rho-dependent termination of transcription?
-The Rho protein contributes to Rho-dependent termination by binding to the RNA at the Rho Utilization Site (RUT site), using ATP to move along the RNA, and catching up with the RNA polymerase. It then sterically hinders or unwinds the DNA-RNA hybrid, causing the release of the RNA transcript and termination of transcription.
What are the differences between prokaryotic and eukaryotic transcription in terms of complexity?
-Eukaryotic transcription is more complex than prokaryotic transcription due to the presence of a larger genome with more genes and non-coding DNA, the necessity of RNA export from the nucleus to the cytoplasm, and the presence of chromatin. Additionally, eukaryotes have three different RNA polymerases, each with their own set of genes to transcribe, and require processing of precursor mRNA, including the removal of introns and addition of exons.
What is the role of the TATA box in eukaryotic transcription?
-The TATA box, also known as the Hogness box, is a DNA sequence element found in the promoter region of many eukaryotic genes. It plays a crucial role in the initiation of transcription by providing a binding site for the TATA-binding protein (TBP) and other transcription factors, which help assemble the transcription pre-initiation complex.
How do enhancers and silencers regulate transcription in eukaryotes?
-Enhancers and silencers are regulatory DNA sequences that can be located at a distance from the promoter and can influence the rate of transcription. Enhancers bind activator proteins that stimulate transcription, while silencers bind repressors that inhibit transcription. They can modulate the transcription machinery's assembly and function, either promoting or suppressing gene expression.
What is the significance of the order in which transcription factors bind during eukaryotic transcription initiation?
-The order in which transcription factors bind is significant as it ensures the proper assembly of the transcription pre-initiation complex. For example, transcription Factor 2D (containing TBP and TAFs) binds the TATA box first, followed by the binding of other factors like 2B, RNA polymerase II, and then 2F and 2H, which together form the complete initiation complex and trigger the start of transcription.
Outlines

هذا القسم متوفر فقط للمشتركين. يرجى الترقية للوصول إلى هذه الميزة.
قم بالترقية الآنMindmap

هذا القسم متوفر فقط للمشتركين. يرجى الترقية للوصول إلى هذه الميزة.
قم بالترقية الآنKeywords

هذا القسم متوفر فقط للمشتركين. يرجى الترقية للوصول إلى هذه الميزة.
قم بالترقية الآنHighlights

هذا القسم متوفر فقط للمشتركين. يرجى الترقية للوصول إلى هذه الميزة.
قم بالترقية الآنTranscripts

هذا القسم متوفر فقط للمشتركين. يرجى الترقية للوصول إلى هذه الميزة.
قم بالترقية الآن5.0 / 5 (0 votes)